What are neutrophils?
Neutrophils are cells of the immune system. They are the most abundant immune cell in human blood. They serve as first responders, quickly migrating to sites of infection or tissue injury. They help to clear the offending agent and attract other cells, as part of a process called inflammation. People who do not have enough neutrophils, or whose neutrophils fail to function properly, are susceptible to infections by bacteria or fungi. People with overactive neutrophils suffer from chronic inflammatory diseases. Neutrophils have also been found to play important roles in the clearance of cancer cells and in autoimmune diseases.
What is the neutrophil transcriptome?
Genes are encoded by DNA. When a gene is expressed, its DNA sequence is read into RNA, in a process known as transcription. The transcriptome is the entire set of genes that are expressed in a given cell or cell type. Activation or repression of gene expression plays a central role in the regulation of cellular function. For many years, neutrophils were believed to be relatively inactive transcriptionally, because they tend to have lower RNA concentrations per cell than other cell types. More recently, neutrophils have been found to express a wide range of genes and to be transcriptionally highly sensitive to environmental stimuli. This growing interest in the neutrophil transcriptome was the motivation for creating NeutGX.
What is NeutGX?
NeutGX is a web application that allows investigators to explore the transcriptome of human neutrophils. The goal is to offer a simple, mobile-friendly tool that can offer quick answers to the following questions: Is my gene of interest expressed in human neutrophils? How variable is its level of expression among healthy human subjects? Are there differences in the expression of my gene of interest between males and females?
How do I use NeutGX?
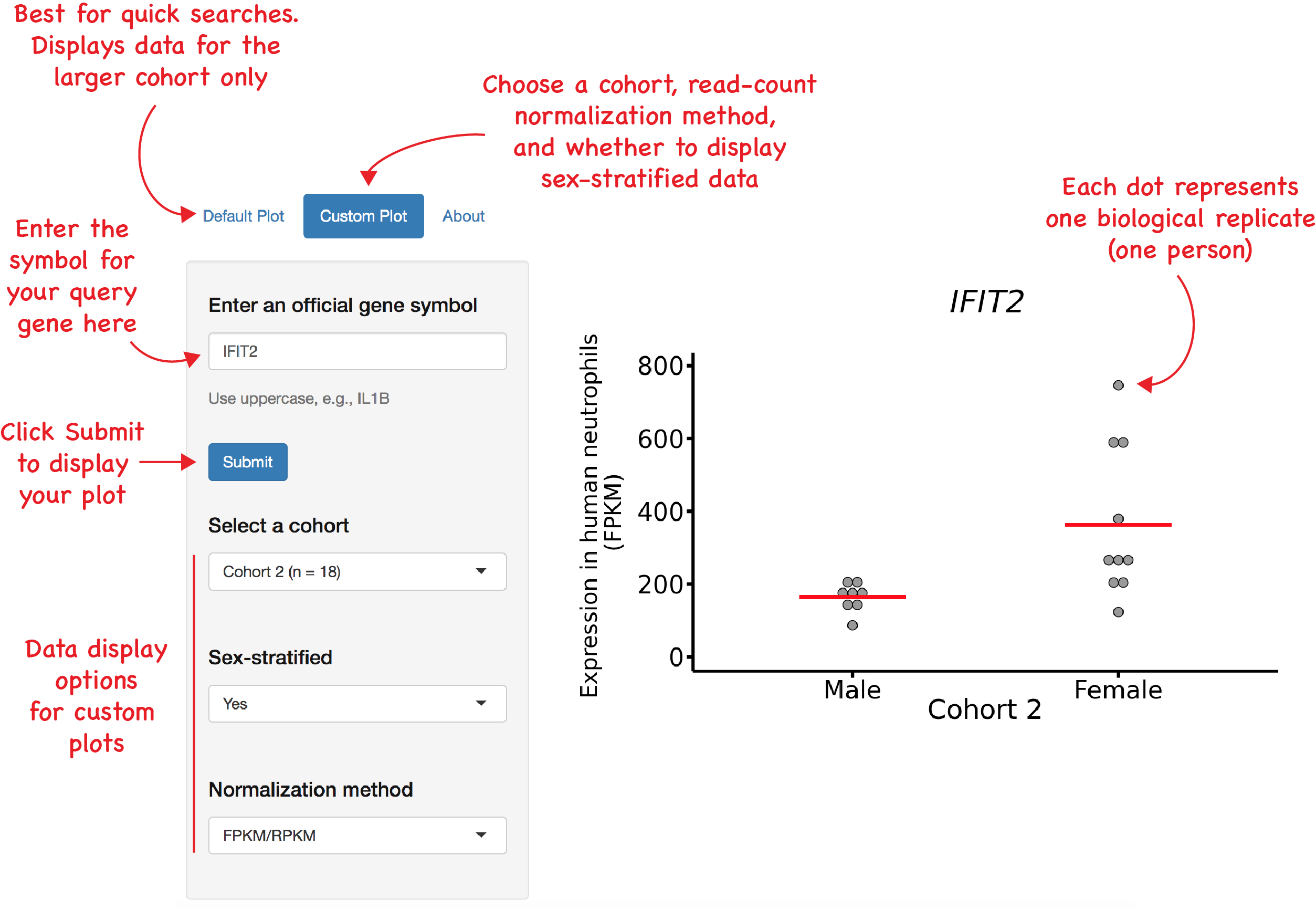
The NeutGX user interface is designed to be simple and self-explanatory. A visual guide is in the diagram below:
Who developed NeutGX?
NeutGX is a joint effort by the Franco and Kaplan labs at the National Institute of Arthritis and Musculoskeletal and Skin Diseases, and the Bioinformatics and Computational Biosciences Branch at the National Institute of Allergy and Infectious Diseases, both at the U.S. National Institutes of Health.
How can I cite NeutGX?
If the neutrophil gene expression data from NeutGX helps inform your research, please cite the PNAS paper in which we first described NeutGX:
Gupta et al. Sex differences in neutrophil biology modulate response to type I interferons and immunometabolism. Proc Natl Acad Sci USA. 2020. doi: 10.1073/pnas.2003603117. PMID: 32601182.
Is it OK for me to copy or save a plot from NeutGX?
Yes. You can right-click on a plot generated by NeutGX and copy or save it for personal use or for academic presentations. We do not add the NeutGX name or logo to the plots, to allow users to employ the images in the manner they see fit, but we do ask that you please give due credit to the NeutGX application when you display images obtained from it. If you would like to use an image from NeutGX in an online or print publication of any kind (including, but not limited to, peer reviewed manuscripts or marketing materials), please contact us first.
Where do the data for NeutGX come from?
The neutrophil gene expression data displayed by NeutGX is from bulk RNA sequencing (RNA-seq) experiments performed on highly pure neutrophils obtained from peripheral blood of healthy human volunteers. Two independent cohorts, with a total of 32 biological replicates (unrelated subjects) were studied separately. Cohort 1 was composed of 14 biological replicates: 7 males and 7 females. Cohort 2 was composed of 18 biological replicates: 8 males and 10 females. Neutrophil purity was documented by flow cytometry. RNA was obtained from purified neutrophils and short-read RNA-seq was performed. Additional details of the methods are in our PNAS paper.
What can I do if my gene of interest is not displayed by NeutGX?
1. Please be sure that you are entering an official gene symbol approved by the HUGO Gene Nomenclature Committee (HGNC).
2. Most genes have had one or more aliases over time; please check the Entrez Gene or
HGNC websites to ensure that you are using is the currently accepted symbol for your gene of interest.
3. Please be sure that your gene symbol is written in uppercase (e.g., IL1B or TNF).
Italics are not necessary.
4. If you have documented that you are entering an official, currently accepted HGNC
gene symbol written in uppercase, and it is not being displayed by NeutGX, please contact us.
Are the raw data publicly available?
Yes. Data are available through GEO with accessions GSE145231 (Cohort 1) and GSE145033 (Cohort 2). The GEO entries include links to the raw data in SRA